Pulling in iNaturalist Data (Very Rough Draft UI)
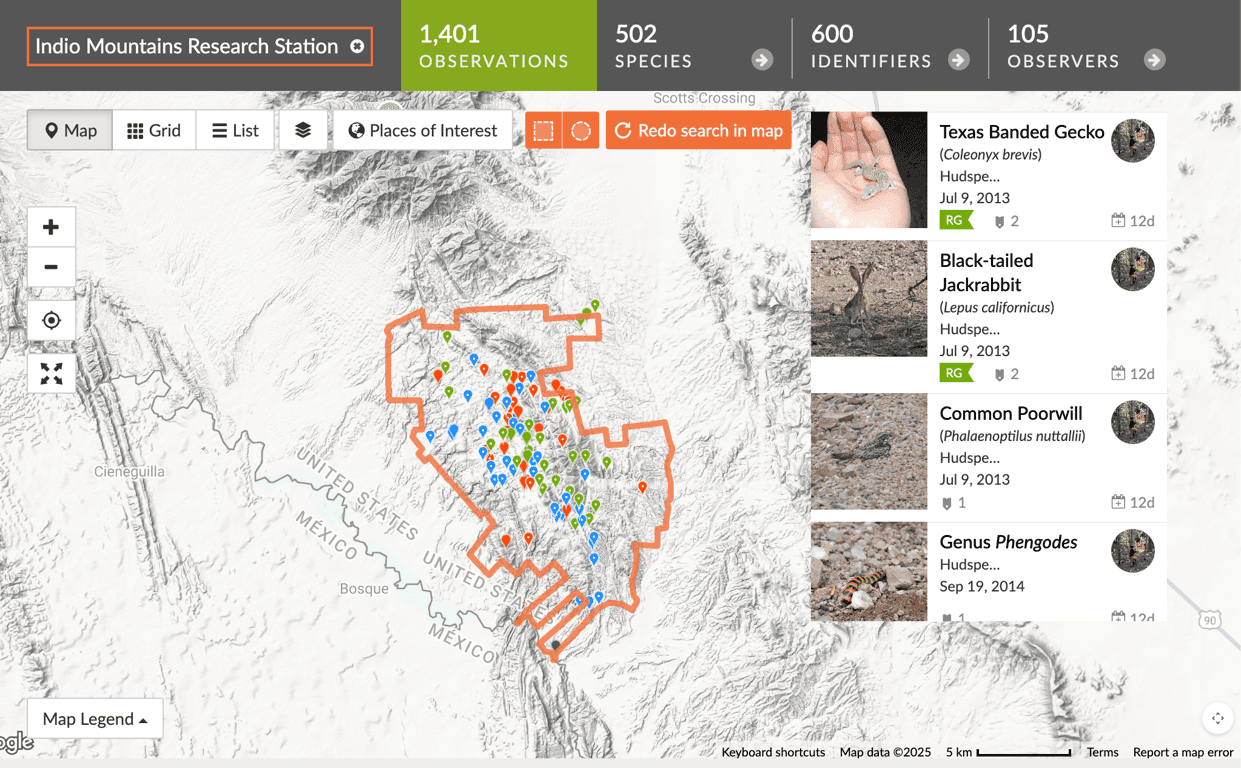
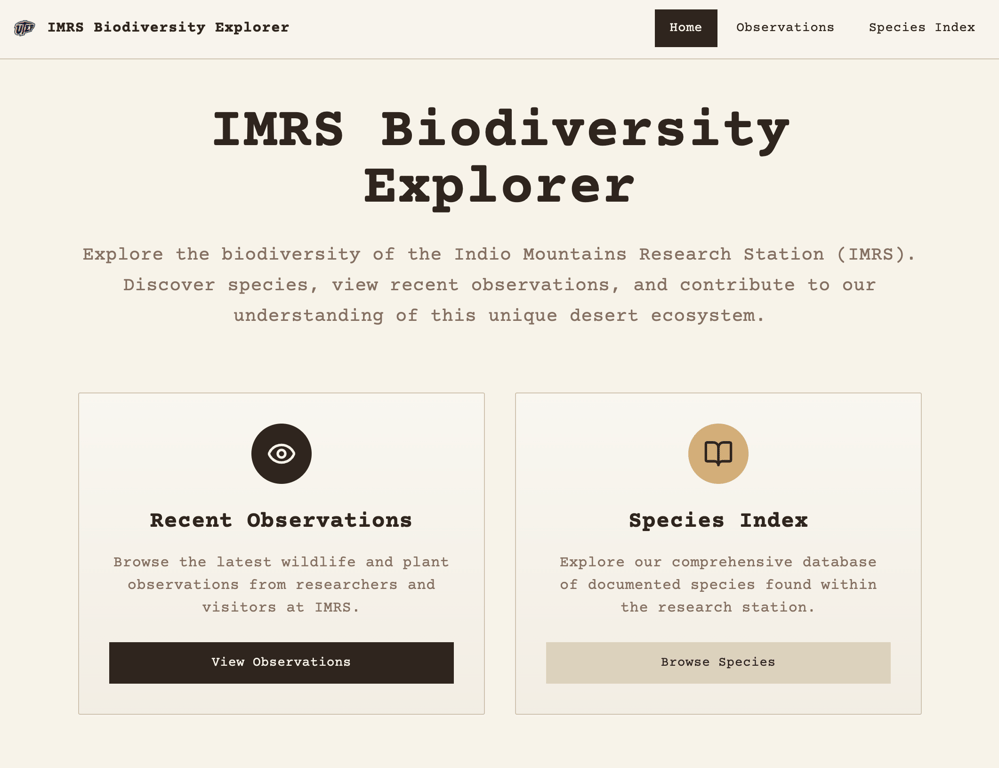
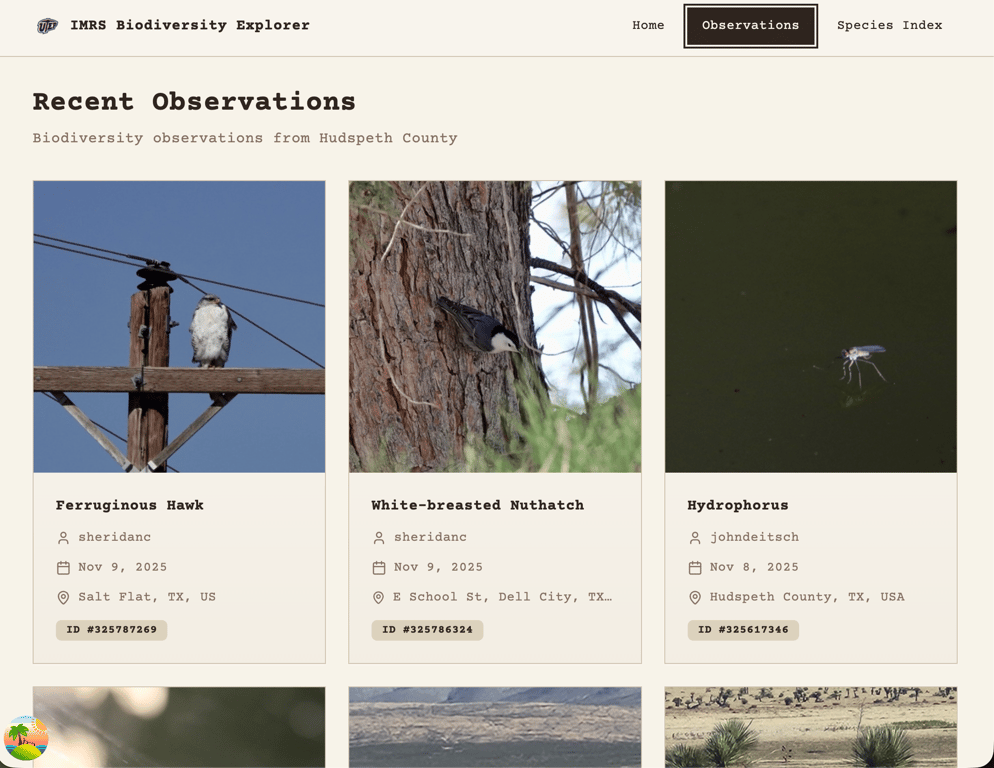
On this installment of the IMRS Project Logs, I am unveiling my very basic UI that is pulling in actual data from iNaturalist (Hudspeth County observations) plus 10 dummy species from a JSON file. Nothing is optimized, I haven’t separated the logic that’s making the calls, the JSON calls are duplicated, the iNaturalist data is capped at the 20 most recent observations, and I still haven’t narrowed it down to Indio Mountains Research Station observations.
 Aside from refactoring these files and making my calls with Tan Stack Query, next on the research list is figuring out how to add a geofence delineating IMRS property and somehow use the iNaturalist API to cross reference the coordinates of an observation in Hudspeth County and determine if it's inside or outside the IMRS geofence. Sounds kind of janky. There's gotta be a better way!
Aside from refactoring these files and making my calls with Tan Stack Query, next on the research list is figuring out how to add a geofence delineating IMRS property and somehow use the iNaturalist API to cross reference the coordinates of an observation in Hudspeth County and determine if it's inside or outside the IMRS geofence. Sounds kind of janky. There's gotta be a better way!
 Figuring Out the SQL Data Structure
Figuring Out the SQL Data Structure
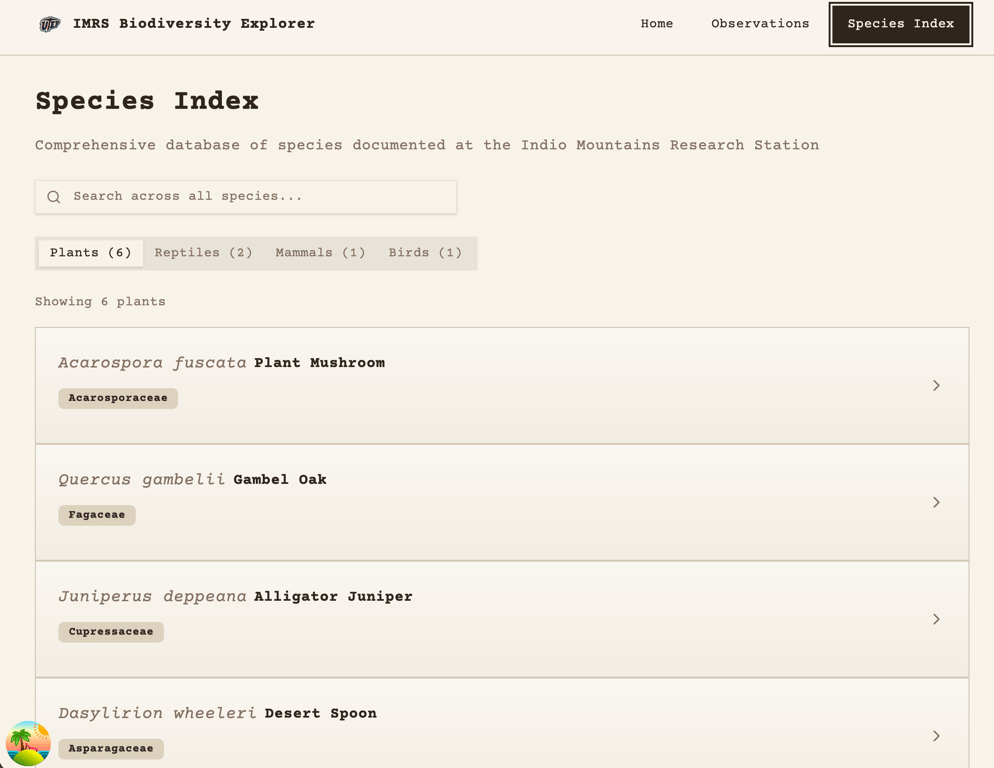
As for the SQL data structure, I kept flip flopping on how I wanted to set up the table(s). Initially I thought I would be storing about 5,000 species but it turns out to only be about 1,200 so I'm going to call my database "small". Columns will include taxonomic ranks (Kingdom → Phylum → Order → Family → Genus → Species), plus notes, collectors’ field numbers, authorship, etc. Growth is minimal with maybe 1–2 new species per year, so the dataset is pretty stable.
My conundrum was deciding whether or not to keep it as one wide table or break it into multiple related tables, a more normalized SQL DB. The data is hierarchical, which suggests reducing duplicate taxonomic levels. I got this idea from the BioSQL Schema Overview, which describes SQL DBs holding genomic data that can be around 1.5 GB for just one full human sequence. I’m not even going to guess how many rows that is. Decision no. 1 – single wide table for a small database.
Choosing a Relational Database
Now which relational db will I actually use? I thought of Supabase because the app will potentially have authentication down the road for admins to add/edit/delete species in the database, but the free tier didn't seem like it would be enough memory for my needs. Then I stumbled upon an oldie but goodie, SQLite! I know it's super popular and used in pretty much all mobile apps. Since my data is small and pretty stable, I thought this would be the perfect fit. Decision no. 2 - SQLite as the database engine.
Hosting the SQLite Database
Of course, that immediately raised the question of where to host it. My initial search made me believe that Vercel did not have the ability to host a SQLite DB, so I started looking at Heroku, Turso, AWS, DigitalOcean, and other relatively cheap hosting options but my goal budget is $0, so I had to keep looking. Turns out I didn’t dig very deep the first time, and I eventually found out Vercel has partnered with Turso to host SQLite databases. Five GB of storage and 500M monthly reads, all for free?!?! Sign me up! Decision no. 3 – cloud hosting on Turso connected through Vercel.
Next Steps: Converting the PDF and Exploring TanStack Query
Now that I have these major DB decisions made, I need to convert the PDF into CSV data to import into SQLite. I’ll definitely leverage AI to do the grunt work of formatting the text into columns. We’ll see how that goes. While I’m getting the data situated, next steps are to look into TanStack Query and take advantage of all the cool features it offers out of the box. Stay tuned!
If you want to poke around the very rough-but-functional UI, here's the latest deployed version: