Building the IMRS Biodiversity Explorer
For the past couple of weeks, I have been slowly building out the IMRS Biodiversity Explorer web app. I decided on TanStack Start mainly to test out a new framework, but also to lean into tanstack-query and tanstack-router, two packages I had not worked with yet but heard great things about!
The MVP for the app has two goals. 1. Fetch and display recent iNaturalist observation data of flora and fauna found on IMRS and 2. Digitize the current Natural Resources PDF (species index) so users can search, filter, and sort species easily. I have to keep reminding myself of these two goals because the scope creep is real!
The First Blocker: Too Much Data
In my last iteration, I pulled in data from iNaturalist and set the location to Hudspeth County, which surrounds IMRS. While this worked, it meant the fetched data included observations beyond IMRS.
My initial idea was to add a GeoJSON boundary and write logic to check whether each observation occurred inside or outside the boundary. Although this is probably possible, it felt cumbersome, and I thought there had to be a better way.
Documentation Deep Dive #1
After digging into the docs, I found that iNaturalist offers a way to add a place. That meant all I would need to change in my fetch query was the place_id.
Problem solved… almost.
One of the stipulations for being granted access to add a place was a blocker:
In order to create a new Place, your account must have a confirmed email address and made at least 50 verifiable observations.
I made my original account using a school email back in 2013 and since I lost access to that email, I made a new account in 2016. I could have sworn I had added observations back then but a lot can happen in 9 years (apparently including forgetting to log a single one). Anywho, I knew what I had to do — go down memory lane.
Going Down Memory Lane
I spent two days going through thousands of photos from my time as an undergraduate and graduate field researcher. The photos were on an external hard drive that kept overheating and it would unexpectedly eject itself every so often which made me anxious because I didn't want any of my files to get corrupted. Going through those images brought back great memories of bug bites, fun hikes and herping at night with the best company! I was fortunate to meet so many people through Field Biology classes, summer research internships, and visits from other universities and departments at UTEP. I spent countless nights under the stars, far enough from light pollution to see an arm of the Milky Way stretch across the sky on a clear night. Using a sweep net on vegetation was always a fun time because you never know what you would catch. I was pretty much a real life Pokemon collector. I gained a deep appreciation for the Chihuahuan Desert and loved seeing it come alive through the seasons. I just might have to plan a trip out there soon!
 Adding IMRS as a Place in iNaturalist
Adding IMRS as a Place in iNaturalist
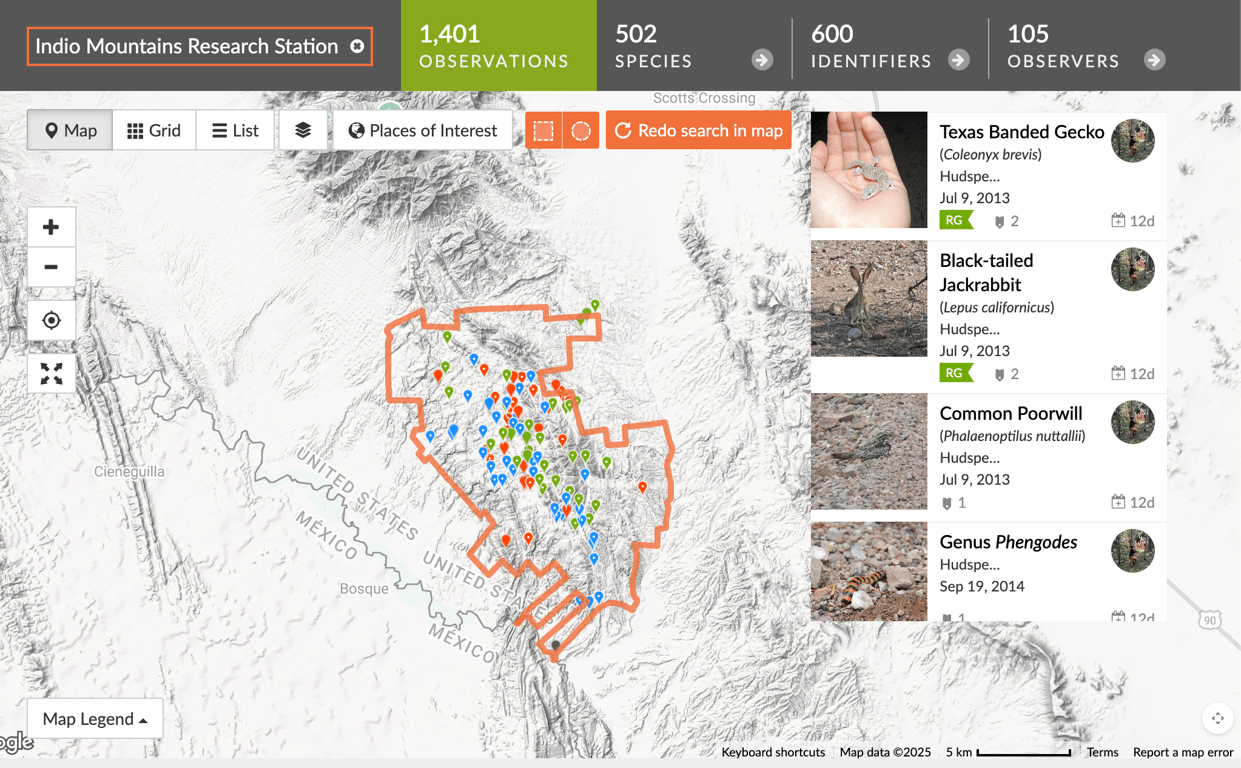
After a lot of patience, I finally hit my 50 observations. I made sure to only include images whose coordinates I was 100% confident in. The next step was actually adding the place to iNaturalist. I reached out to the current director of IMRS, who provided me with a KMZ file of the property boundary. I converted it to a KML file via Google Earth, uploaded it to iNaturalist, and played the waiting game.
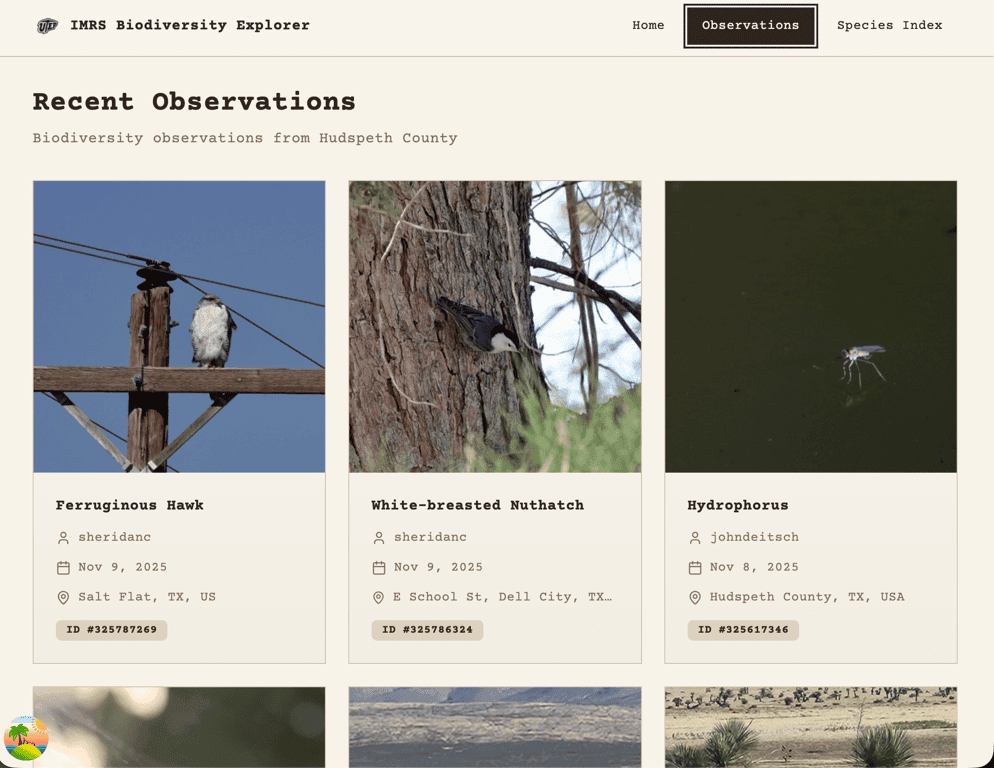
 Fetching Observations (The Clean Way)
Fetching Observations (The Clean Way)
Once Indio Mountains Research Station was added to iNaturalist, only a few hours later over 1,400 observations were populated! I officially had a new place_id to query.
This allowed me to completely bypass the custom boundary logic I had originally planned! What a time saver!
I used React Query to fetch the data and implemented useInfiniteQuery so users could scroll indefinitely and have observations continue loading. The first query is server-side rendered, seeding the initial observation cards, and subsequent pages are fetched on the client. iNaturalist made this part easy too because the data fetched was already paginated!
I also realized it's not good practice to set the staleTime or gcTime for the query made in the route loader. I like to think of it as routes own data, components render data. Since this data does not change often, I set:
staleTime to 30 days
gcTime to 60 days
I will probably adjust this later, but it felt like a reasonable starting point.
I am using shadcn/ui as my component library and leveraged the Skeleton component as a placeholder for observation cards while data is loading.
Open Questions & Architecture Decisions
Right now, the observations query lives directly in the router loader (which I think is pretty nifty). However, I may move this logic into its own api/observations endpoint in the future.
The species details page also pulls in iNaturalist data, but it only fetches four recent observations, and they aren’t necessarily from IMRS. This leaves me with another decision to weigh in on:
Should I prioritize IMRS observations first, and only fall back to global observations if none exist?
Or should I just keep the current global observations?
I still need to brainstorm this and decide whether a reusable api/observations endpoint makes more sense long-term.
Documentation Deep Dive #2
Since my last blog post, I also updated the Species Index page. Although the current site is rendering real data from Turso, I was stumped for a while as to why fetching data was giving me such a hard time. I was able to query data locally when it just lived as JSON in my data.ts file but I needed a real backend endpoint to query the DB. I made my api route and tried including logic to only fetch locally and it didn't work. I couldn’t figure out why my api/species endpoint wasn’t being recognized.
After a couple hours of debugging, I finally figured it out........ I misnamed the file.
Tan Stack is a very opinionated framework but for good reason. The answer was right there in the docs! instead of /routes/api/species.ts I had named my file /routes/api.species.ts No wonder it couldn't reach the backend, the endpoint didn't exist! I’m proud to say that the Species Index page is now rendering real SQLite data hosted on Turso. It’s still dummy data, but the api/species endpoint is working!
The Current Blocker: Data Sanitization
I ran into a snag when exporting real species data as a CSV and importing it into SQLite. Many of the notes and records contain commas, which caused the number of values to mismatch the actual table columns on import. I need to sit down, maybe write some Python scripts and get this baby going!The last blocker is a good reminder of a pattern I keep seeing in this project: almost every "hard" problem so far hasn't been solved with overly complicated engineering, just slowing down and actually reading the documentation. Time to sit down, read some docs and blogs because I know I'm not the first one to run into this problem.
Once the data is sanitized, the MVP will be complete! Here's your friendly reminder to always start with the docs and reread!